Pedersen Lab
Principal Investigator: Irene Pedersen, PhD
RESEARCH
The research interest of the Irene Munk Pedersen (IMP) laboratory is based on the recent discovery of microRNAs (miRs), which has revolutionized our understanding of gene control. miRs play key roles in controlling development, stem cell fates and differentiation, and mutations in human miR genes have been linked to human disease.
The main focus of the lab is to investigate novel roles of specific miRs (identified using anti-miR library screens approaches) in cells and explore how to use RNA molecules (miRs and mRNAs) as tools to regulate/counteract potential disregulation in cancer (Research Program 1) and neurological disorders (Research Program 2). A separate line of research interest is to evaluate miRs function as restriction factors of mobile elements and retrovirus (LINE-1, hERVs and HIV-1) in human cells (Research Program 3).
RESEARCH PROGRAM 1
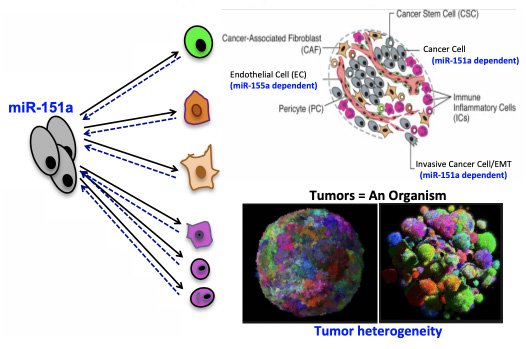
miR-151a’s role in the lung cancer niche
We have previously established that miRs can serve as a novel link between inflammation and tumorigenesis. This work has resulted in a clinical trial testing an anti-miR-155 regimen for patients with hematological disorders. In parallel, we performed studies to evaluate miRs role as it relates to cell-cell contact and hallmarks of tumorigenesis including metastasis and angiogenesis. To this end we have demonstrated that miR-151a function as an onco-miR and that expression is significantly increased in lung cancer specimens relative to normal lung. miR-151a promotes tumor cell growth, partial EMT and tumor-associated angiogenesis. As a logical next step, we have initiated the characterization of genetic and non-genetic heterogeneity of miR-151a in the lung cancer niche (tumor-microenvironment interactions), by taking an scRNA-seq approach.
miR-151a’s role in lung cancer
miR-151a induces partial EMT
miR-151a enhances angiogenesis
miR-151a enhances angiogenesis
RESEARCH PROGRAM 2
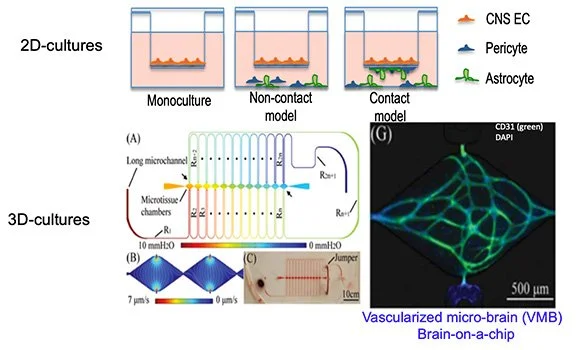
miR-23b’s role in repairing damaged endothelium
The blood-brain barrier (BBB) plays a critical role in safeguarding the homeostasis of the brain’s microenvironment. The barrier is formed by brain endothelial cells (BECs), which are sealed by specialized tight junctions (TJs) and exhibit minimal caveolar-mediated transport (transcytosis), thus preventing the passage of hydrophilic molecules between blood and brain. We recently made the following advances: 1) developed a safe and efficient reprogramming strategy using RNA modulation of cells and 2) performed a novel anti-microRNA (miR) screen in BECs and showed that anti-miR-23b significantly enhances BEC function by enhancing both expression of TJ proteins and formation of a paracellular endothelial barrier. We’re in the process of optimizing protocols to successfully differentiate iPSCs into BECs with mature BBB properties, by taking advantage of the unique properties of anti-miR-23b alone or in the combination with other RNAs. We are exploring the mechanisms by which miR-23b regulates angiogenic and barrier properties of BECs in vitro and will test the ability of anti-miR-23b to prevent or repair the injured BBB in models for neurological disorders (stroke and Alzheimer’s disease), by using a novel gene therapy approach.
RNA-only Reprogramming Strategy
Using anti-miRs and mRNAs as a tool to enhance iPSC-derived BBB EC function
Approaches to study RNA enhanced BBB EC properties
anti-miR gene therapy approach to repair damaged endothelium
RESEARCH PROGRAM 3
miR-128’s function as a restriction factor of mobile elements and retrovirus
Repetitive elements, including LINE-1 (L1), comprise approximately half of the genomes of humans. These elements can potentially destabilize the genome by activation and reintegration into new sites (retrotransposition). In somatic cells, active L1 elements are repressed by distinct molecular mechanisms including DNA methylation and histone modifications to repress transcription. Under conditions of hypomethylation (e.g. tumor cells and in stem cells) additional restriction mechanisms include silencing by miRs. We recently demonstrated that miR-128 represses L1 activity by a dual mechanism by directly binding to a site in the coding region of L1 RNA and targeting required host factors. Interestingly, miR-128 is an interferon inducible gene and as such seems to function as a master regulator of an overlapping network of host co-factors which L1 and HIV-1 are dependent on for efficient replication. We are in the process of characterizing this network of requited host factors.